Prior-guided Prototype Aggregation Learning for Alzheimer's Disease Diagnosis
MICCAI 2025 Conference Poster
Abstract
Alzheimer's Disease (AD) is a progressive neurodegenerative disorder that represents a major global health challenge. Early and accurate diagnosis is crucial for effective intervention and treatment planning. We propose Prior-guided Prototype Aggregation Learning (PPAL), a novel framework that leverages disease-related prior knowledge to enhance AD diagnosis from multi-modal brain imaging data.
Our approach introduces a unique question-and-answer mechanism to extract disease-related concepts from medical knowledge bases. By integrating these semantic priors with visual features through progressive prototype aggregation, PPAL achieves more robust and interpretable disease classification.
The framework includes a slice selection and compression module to identify and integrate critical information from 3D brain scans, and a text-aggregated visual module that progressively refines prototype features using text embeddings. Extensive experiments on the ADNI dataset demonstrate the effectiveness of our approach for AD diagnosis.
Key Contributions
- Prior-guided Learning: Novel integration of medical knowledge priors through a question-and-answer mechanism for disease concept extraction
- Prototype Aggregation: Progressive aggregation of visual features using text embeddings to obtain refined prototype representations
- Slice Selection Module: Intelligent selection and integration of key slice information to enhance critical representations from 3D brain imaging
- Semantic Similarity Classification: Classification based on semantic similarity measurements between prototypes and query samples
- Multi-modal Integration: Effective fusion of visual and textual modalities for improved diagnostic performance
Method Overview
The PPAL framework consists of four main components working in synergy to achieve accurate and interpretable Alzheimer's Disease diagnosis:
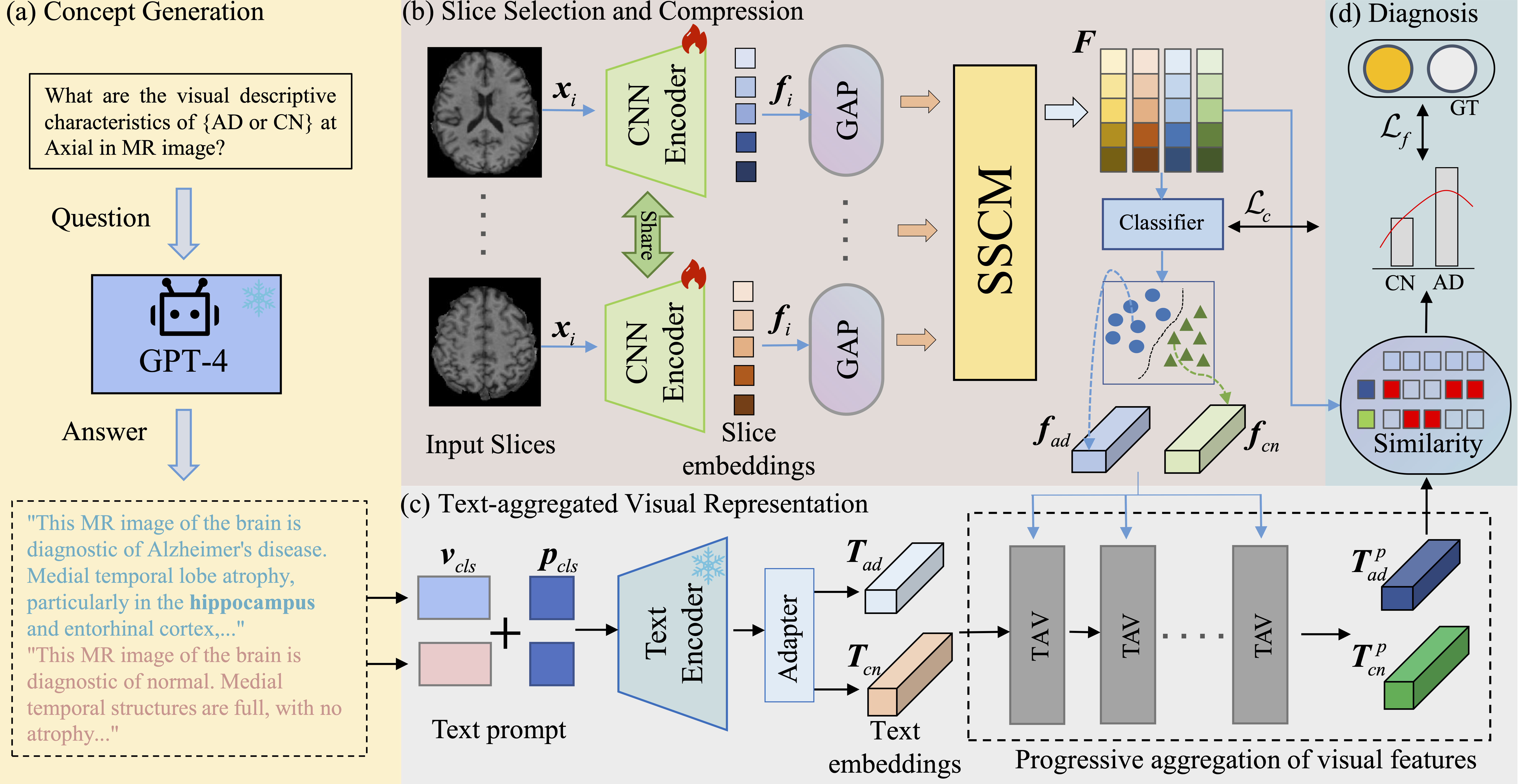
1. Disease Concept Extraction
The first component employs a question-and-answer mechanism to extract disease-related concepts from medical knowledge bases. This approach leverages prior medical knowledge to guide the learning process, ensuring that the model focuses on clinically relevant features associated with Alzheimer's Disease progression.
2. Slice Selection and Compression Module
Given the 3D nature of brain imaging data, it's crucial to identify the most informative slices across different anatomical planes. Our slice selection and compression module intelligently selects and integrates key slice information to enhance critical representations.
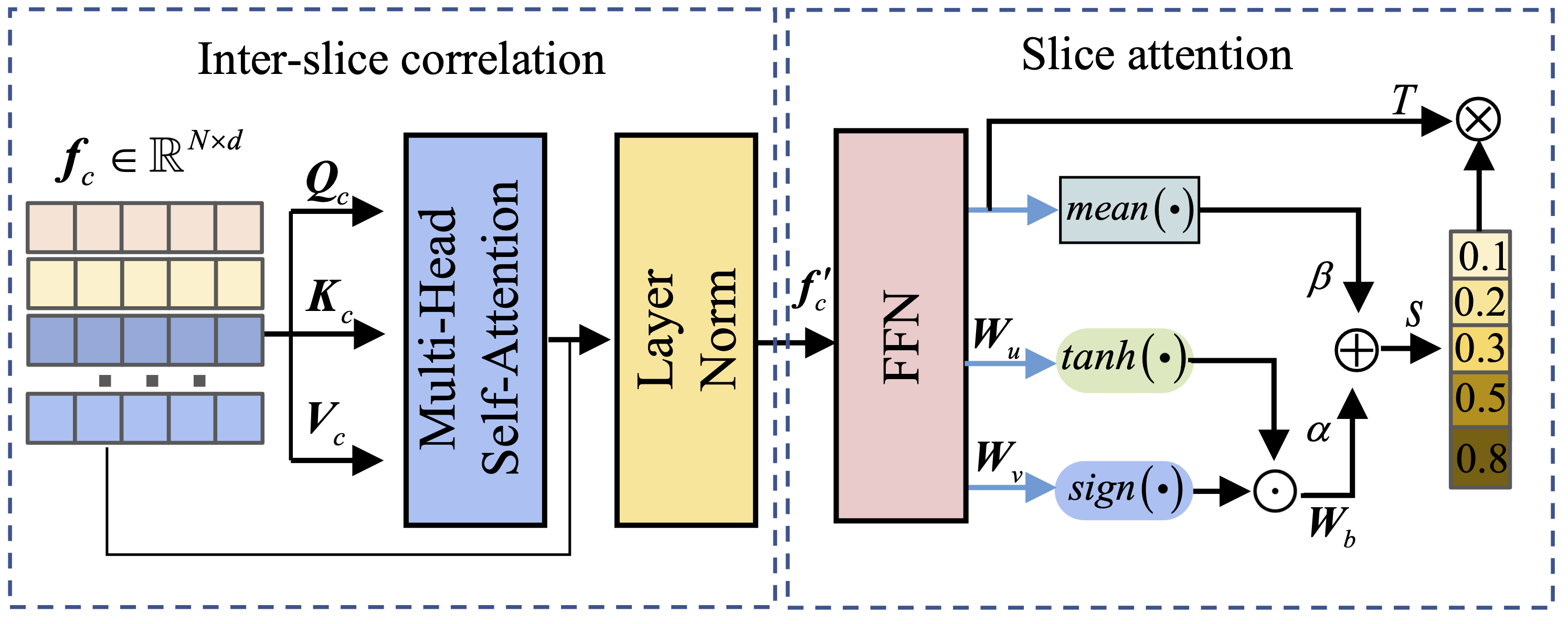
3. Text-Aggregated Visual Module
The core innovation of PPAL lies in the text-aggregated visual module, which progressively aggregates visual features using text embeddings derived from disease-related concepts. This cross-modal fusion enables the model to learn more refined prototype feature representations that capture both visual patterns and semantic disease characteristics.
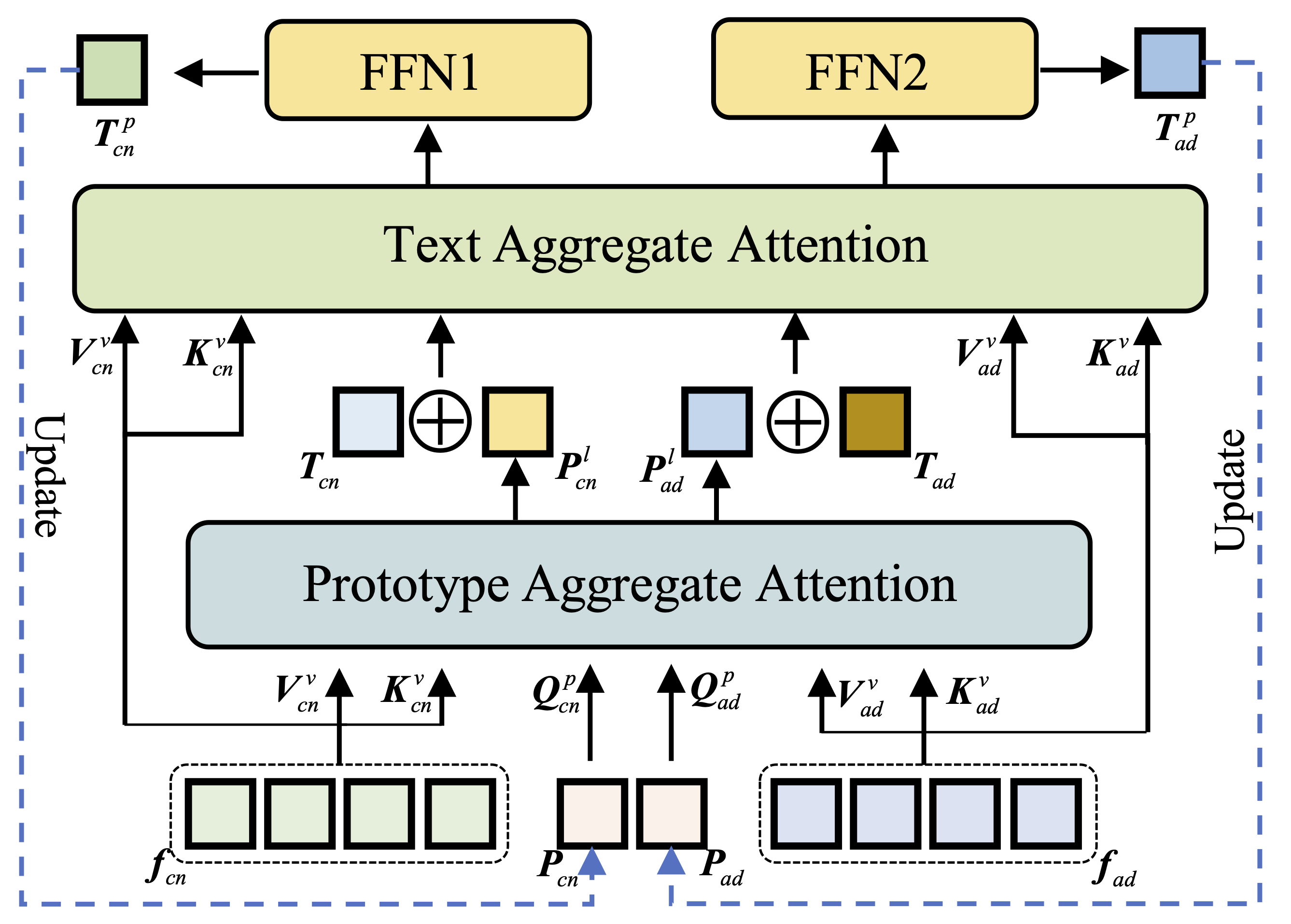
4. Semantic Similarity-Based Classification
Classification is performed based on semantic similarity measurements between the learned prototypes and query samples. This approach provides interpretable predictions by explicitly comparing the similarity of patient scans to disease-specific prototype patterns.
Results & Visualization
We evaluate PPAL on the ADNI (Alzheimer's Disease Neuroimaging Initiative) dataset, which contains multi-modal brain imaging data from patients with various stages of Alzheimer's Disease progression.
Model Interpretability
One of the key advantages of PPAL is its interpretability. The framework provides insights into the decision-making process through:
- Slice Attention Visualization: Shows which anatomical planes and slices the model focuses on for diagnosis
- ROI Attention Maps: Highlights specific brain regions that contribute most to the classification decision
- Prototype Similarity Analysis: Reveals how patient scans relate to learned disease prototypes
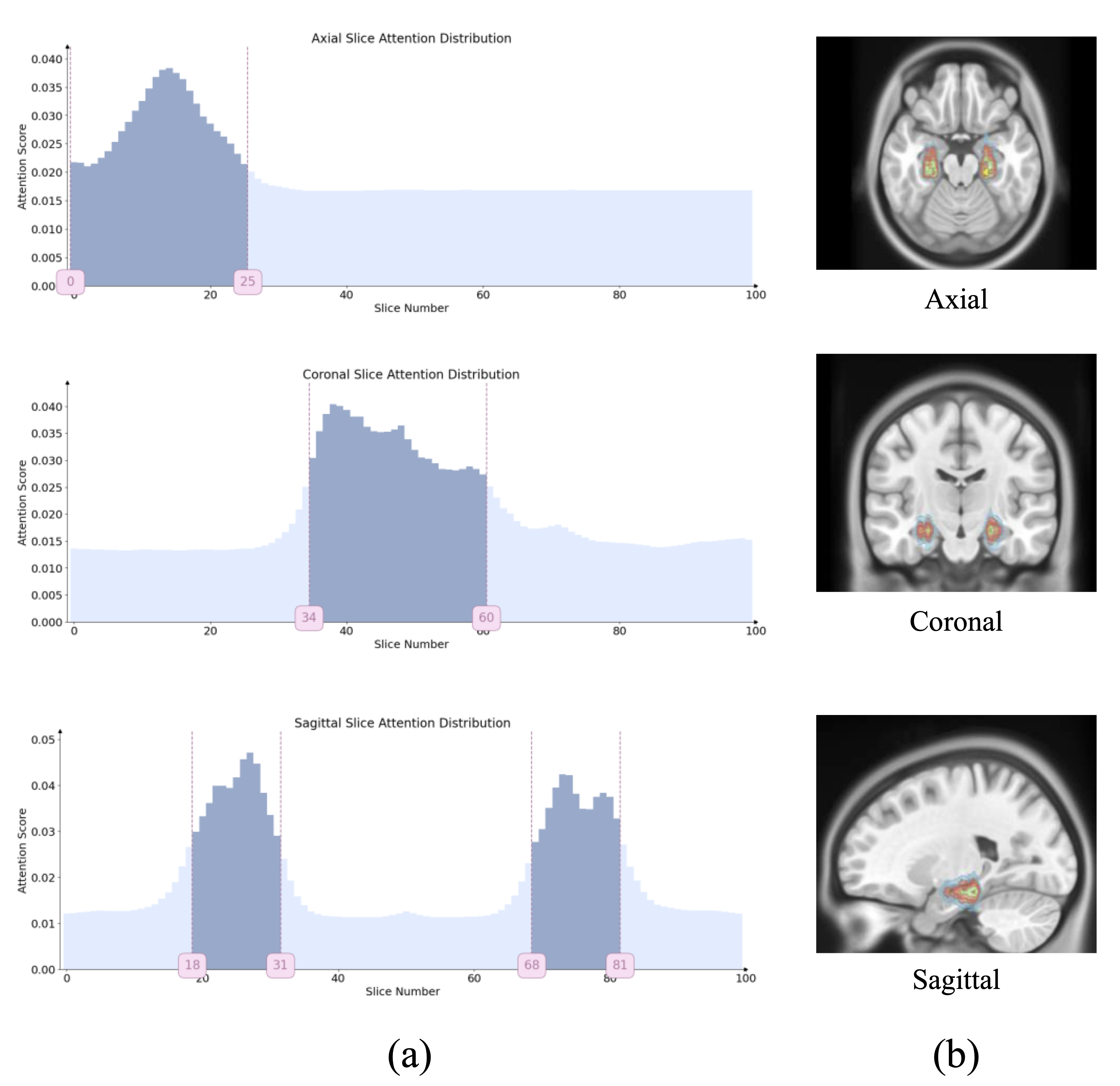
The attention visualizations in Figure 4 demonstrate that PPAL learns to focus on clinically relevant brain regions known to be affected by Alzheimer's Disease, such as the hippocampus, entorhinal cortex, and temporal lobes. This alignment with medical knowledge validates the effectiveness of the prior-guided learning approach.
Dataset & Code
ADNI Dataset
The Alzheimer's Disease Neuroimaging Initiative (ADNI) is a landmark study that has been collecting clinical, imaging, genetic, and biomarker data from thousands of participants. The dataset includes:
- Structural MRI: High-resolution T1-weighted brain imaging across multiple time points
- Clinical Assessments: Comprehensive cognitive and functional evaluations
- Multi-stage Diagnosis: Including normal cognition (CN), mild cognitive impairment (MCI), and Alzheimer's Disease (AD)
Access to the ADNI data requires registration and agreement to data usage terms. Visit the ADNI website for more information.
Code Availability
The implementation of PPAL is available on GitHub. The repository includes:
- Model Architecture: Complete implementation of all PPAL components
- Training Scripts: Code for training on ADNI dataset
- Evaluation Tools: Scripts for model evaluation and visualization
- Pre-trained Models: Checkpoints for reproducing results
Visit the GitHub repository to access the code and documentation.
Impact & Applications
PPAL addresses critical challenges in computer-aided diagnosis of Alzheimer's Disease by combining medical knowledge priors with advanced deep learning techniques. The framework's potential applications include:
- Early Detection: Assisting clinicians in early identification of AD-related brain changes
- Disease Staging: Differentiating between different stages of cognitive decline
- Treatment Planning: Providing interpretable insights to support personalized treatment strategies
- Clinical Research: Enabling large-scale analysis of brain imaging biomarkers in AD studies
- Multi-disease Diagnosis: The framework's architecture can be adapted to other neurodegenerative diseases
The interpretability of PPAL is particularly valuable in clinical settings, where understanding the reasoning behind AI predictions is essential for building trust and facilitating human-AI collaboration in medical decision-making.
Citation
@inproceedings{diao2025ppal,
title={Prior-guided Prototype Aggregation Learning for Alzheimer's Disease Diagnosis},
author={Diao, Yueqin and Fang, Huihui and Yu, Hanyi and Wang, Yuning and Tao, Yaling and Huang, Ziyan and Yeo, Si Yong and Xu, Yanwu},
booktitle={Medical Image Computing and Computer Assisted Intervention -- MICCAI 2025},
pages={483--492},
year={2025},
organization={Springer}
}
Published in MICCAI 2025
Medical Image Computing and Computer Assisted Intervention